How to construct knowledge graphs
In this guide we'll go over the basic ways of constructing a knowledge graph based on unstructured text. The constructured graph can then be used as knowledge base in a RAG application.
⚠️ Security note ⚠️
Constructing knowledge graphs requires executing write access to the database. There are inherent risks in doing this. Make sure that you verify and validate data before importing it. For more on general security best practices, see here.
Architecture
At a high-level, the steps of constructing a knowledge graph from text are:
- Extracting structured information from text: Model is used to extract structured graph information from text.
- Storing into graph database: Storing the extracted structured graph information into a graph database enables downstream RAG applications
Setup
First, get required packages and set environment variables. In this example, we will be using Neo4j graph database.
%pip install --upgrade --quiet langchain langchain-neo4j langchain-openai langchain-experimental neo4j
[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m A new release of pip is available: [0m[31;49m24.0[0m[39;49m -> [0m[32;49m24.3.1[0m
[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m To update, run: [0m[32;49mpip install --upgrade pip[0m
Note: you may need to restart the kernel to use updated packages.
We default to OpenAI models in this guide.
import getpass
import os
os.environ["OPENAI_API_KEY"] = getpass.getpass()
# Uncomment the below to use LangSmith. Not required.
# os.environ["LANGCHAIN_API_KEY"] = getpass.getpass()
# os.environ["LANGCHAIN_TRACING_V2"] = "true"
········
Next, we need to define Neo4j credentials and connection. Follow these installation steps to set up a Neo4j database.
import os
from langchain_neo4j import Neo4jGraph
os.environ["NEO4J_URI"] = "bolt://localhost:7687"
os.environ["NEO4J_USERNAME"] = "neo4j"
os.environ["NEO4J_PASSWORD"] = "password"
graph = Neo4jGraph(refresh_schema=False)
LLM Graph Transformer
Extracting graph data from text enables the transformation of unstructured information into structured formats, facilitating deeper insights and more efficient navigation through complex relationships and patterns. The LLMGraphTransformer converts text documents into structured graph documents by leveraging a LLM to parse and categorize entities and their relationships. The selection of the LLM model significantly influences the output by determining the accuracy and nuance of the extracted graph data.
import os
from langchain_experimental.graph_transformers import LLMGraphTransformer
from langchain_openai import ChatOpenAI
llm = ChatOpenAI(temperature=0, model_name="gpt-4-turbo")
llm_transformer = LLMGraphTransformer(llm=llm)
Now we can pass in example text and examine the results.
from langchain_core.documents import Document
text = """
Marie Curie, born in 1867, was a Polish and naturalised-French physicist and chemist who conducted pioneering research on radioactivity.
She was the first woman to win a Nobel Prize, the first person to win a Nobel Prize twice, and the only person to win a Nobel Prize in two scientific fields.
Her husband, Pierre Curie, was a co-winner of her first Nobel Prize, making them the first-ever married couple to win the Nobel Prize and launching the Curie family legacy of five Nobel Prizes.
She was, in 1906, the first woman to become a professor at the University of Paris.
"""
documents = [Document(page_content=text)]
graph_documents = llm_transformer.convert_to_graph_documents(documents)
print(f"Nodes:{graph_documents[0].nodes}")
print(f"Relationships:{graph_documents[0].relationships}")
Nodes:[Node(id='Marie Curie', type='Person', properties={}), Node(id='Pierre Curie', type='Person', properties={}), Node(id='University Of Paris', type='Organization', properties={})]
Relationships:[Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='Pierre Curie', type='Person', properties={}), type='MARRIED', properties={}), Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='University Of Paris', type='Organization', properties={}), type='PROFESSOR', properties={})]
Examine the following image to better grasp the structure of the generated knowledge graph.
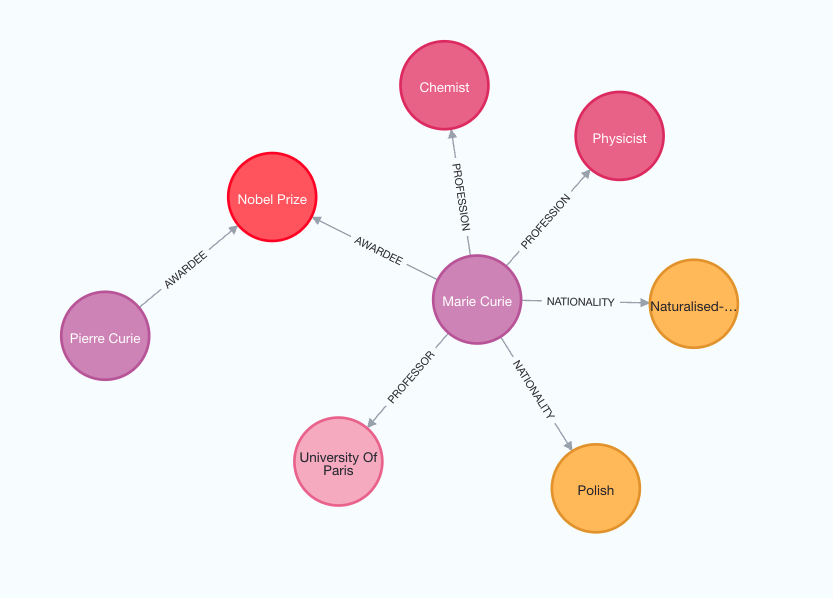
Note that the graph construction process is non-deterministic since we are using LLM. Therefore, you might get slightly different results on each execution.
Additionally, you have the flexibility to define specific types of nodes and relationships for extraction according to your requirements.
llm_transformer_filtered = LLMGraphTransformer(
llm=llm,
allowed_nodes=["Person", "Country", "Organization"],
allowed_relationships=["NATIONALITY", "LOCATED_IN", "WORKED_AT", "SPOUSE"],
)
graph_documents_filtered = llm_transformer_filtered.convert_to_graph_documents(
documents
)
print(f"Nodes:{graph_documents_filtered[0].nodes}")
print(f"Relationships:{graph_documents_filtered[0].relationships}")
Nodes:[Node(id='Marie Curie', type='Person', properties={}), Node(id='Pierre Curie', type='Person', properties={}), Node(id='University Of Paris', type='Organization', properties={})]
Relationships:[Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='Pierre Curie', type='Person', properties={}), type='SPOUSE', properties={}), Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='University Of Paris', type='Organization', properties={}), type='WORKED_AT', properties={})]
To define the graph schema more precisely, consider using a three-tuple approach for relationships. In this approach, each tuple consists of three elements: the source node, the relationship type, and the target node.
allowed_relationships = [
("Person", "SPOUSE", "Person"),
("Person", "NATIONALITY", "Country"),
("Person", "WORKED_AT", "Organization"),
]
llm_transformer_tuple = LLMGraphTransformer(
llm=llm,
allowed_nodes=["Person", "Country", "Organization"],
allowed_relationships=allowed_relationships,
)
graph_documents_filtered = llm_transformer_tuple.convert_to_graph_documents(documents)
print(f"Nodes:{graph_documents_filtered[0].nodes}")
print(f"Relationships:{graph_documents_filtered[0].relationships}")
Nodes:[Node(id='Marie Curie', type='Person', properties={}), Node(id='Pierre Curie', type='Person', properties={}), Node(id='University Of Paris', type='Organization', properties={})]
Relationships:[Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='Pierre Curie', type='Person', properties={}), type='SPOUSE', properties={}), Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='University Of Paris', type='Organization', properties={}), type='WORKED_AT', properties={})]
For a better understanding of the generated graph, we can again visualize it.
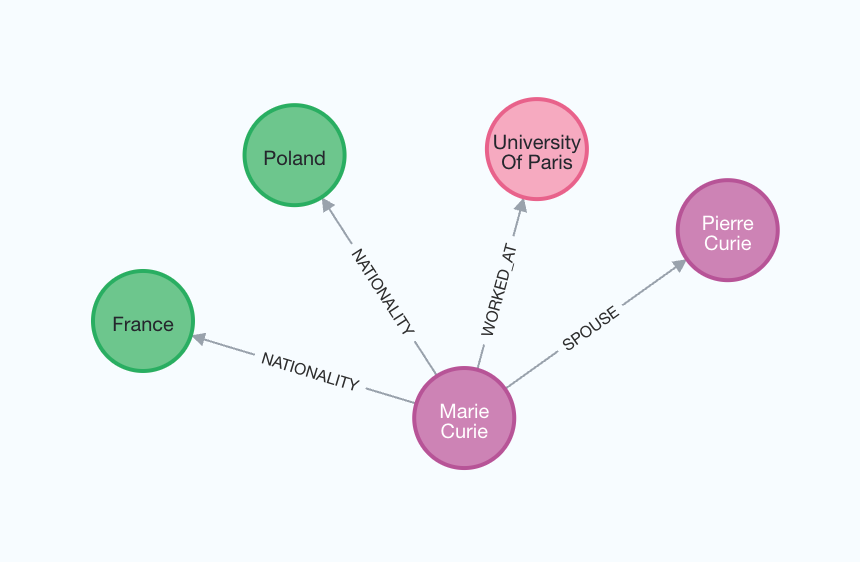
The node_properties parameter enables the extraction of node properties, allowing the creation of a more detailed graph.
When set to True, LLM autonomously identifies and extracts relevant node properties.
Conversely, if node_properties is defined as a list of strings, the LLM selectively retrieves only the specified properties from the text.
llm_transformer_props = LLMGraphTransformer(
llm=llm,
allowed_nodes=["Person", "Country", "Organization"],
allowed_relationships=["NATIONALITY", "LOCATED_IN", "WORKED_AT", "SPOUSE"],
node_properties=["born_year"],
)
graph_documents_props = llm_transformer_props.convert_to_graph_documents(documents)
print(f"Nodes:{graph_documents_props[0].nodes}")
print(f"Relationships:{graph_documents_props[0].relationships}")
Nodes:[Node(id='Marie Curie', type='Person', properties={'born_year': '1867'}), Node(id='Pierre Curie', type='Person', properties={}), Node(id='University Of Paris', type='Organization', properties={}), Node(id='Poland', type='Country', properties={}), Node(id='France', type='Country', properties={})]
Relationships:[Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='Poland', type='Country', properties={}), type='NATIONALITY', properties={}), Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='France', type='Country', properties={}), type='NATIONALITY', properties={}), Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='Pierre Curie', type='Person', properties={}), type='SPOUSE', properties={}), Relationship(source=Node(id='Marie Curie', type='Person', properties={}), target=Node(id='University Of Paris', type='Organization', properties={}), type='WORKED_AT', properties={})]
Storing to graph database
The generated graph documents can be stored to a graph database using the add_graph_documents method.
graph.add_graph_documents(graph_documents_props)
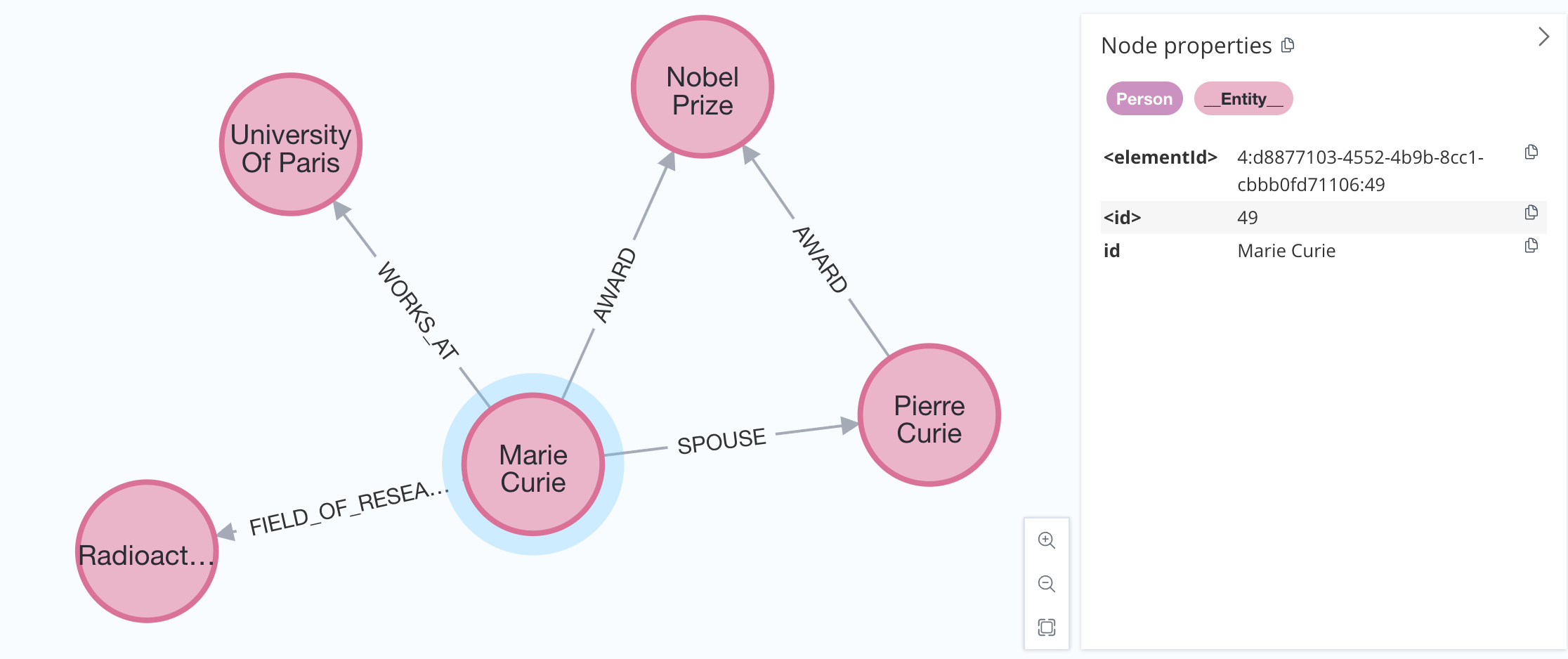
Most graph databases support indexes to optimize data import and retrieval. Since we might not know all the node labels in advance, we can handle this by adding a secondary base label to each node using the baseEntityLabel parameter.
graph.add_graph_documents(graph_documents, baseEntityLabel=True)
Results will look like:
The final option is to also import the source documents for the extracted nodes and relationships. This approach lets us track which documents each entity appeared in.
graph.add_graph_documents(graph_documents, include_source=True)
Graph will have the following structure:
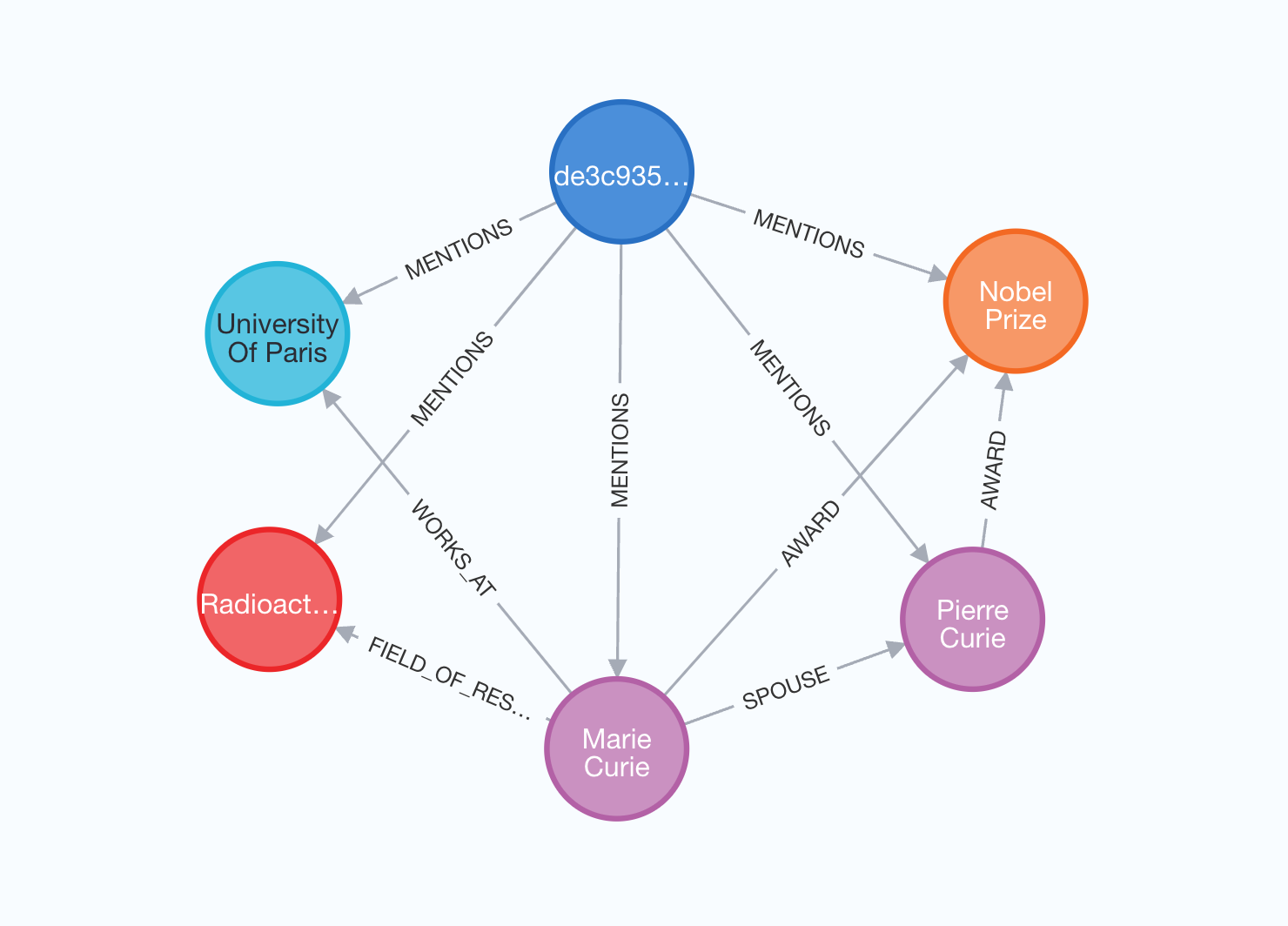
In this visualization, the source document is highlighted in blue, with all entities extracted from it connected by MENTIONS relationships.